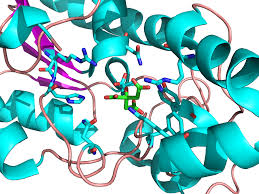
Phosphoglycerate mutase Enzyme
Order Instructions:
ENZYME: Phosphoglycerate mutase
The purpose of this assignment is to encourage you to integrate lecture material on protein structure and enzyme function, with the biochemical and biological role of a key metabolic enzyme. The listed proteins have been chosen because they play an important role in carbohydrate metabolism.
You must research the following information about your protein:
Structure: is it a globular, membrane or structural protein; include a figure showing a molecular model of your protein; describe its structure including secondary structures, any domains, quaternary structure, any prosthetic groups; include the molecular weight and isoelectric point of your protein.
Function: Write an equation for the reaction catalysed by your enzyme; in what pathway does your enzyme participate. In what organisms, organs, tissues, cells, intracellular compartment is the enzyme and pathway found? What is the metabolic purpose of the pathway? What is the importance of your enzyme in that pathway (e.g. regulation, thermodynamics)? Relate the biochemical function of your enzyme to practical information such as clinical data.
Relationship between structure and function: describe any features which are particularly important for catalytic function such as specific amino acids with a key function, the 3 dimensional structure of the active site or binding site. Is your enzyme soluble or membrane protein and how does this relate to its function? What role is played by any co-factors? Is your enzyme regulated? Describe any changes to structure that occur during regulation explaining how the structure affects enzyme activity.
Pitfalls to avoid:
Choose ONE organism as your main focus; the enzymes in the list are found in many different organisms but there will often be major differences between enzymes in humans and E. coli.Sometimes information on one organism may be limited so it may be necessary to have a figure of e.g. a pig enzyme if you are mainly discussing the human version; mixing up info on human and yeast will result in loss of marks.
Be aware that some enzymes are present in several isoforms, do not get these confused.
SAMPLE ANSWER
Question 1
Phophoglycerate mutase (PGM) is a globular protein common in glycolytic activities. The secondary structure is an alpha/beta protein with three main layers as comprising of an alpha/beta/alpha. Additionally, it has a beta sheet comprising of six strands arranged in a specific order with strand 5 occurring as an antiparallel to the rest of the strands. The quaternary structure is made of two identical subunits hence can be categorized as a homodimer. The realative molecular mass of dimers is 55,000 -61,000 kDa. The quaternary is same as the primary, secondary and tertiary structure on the basis of active sites. However, the PGM is has isozymes, which is several variations existing. The type of an enzyme isozyme catalyzing a reaction depends on the tissue where the enzyme is active (Fraser, Kvaratskhelia & White, 1999).
There are two forms of the glycolytic phosphoglycerate mutase (PGM) which are evolutionary unrelated. They occur as non-homologous isofunctional(NISE), each with an independent 3D structure. The first is the co-factor dependent PGM (dPGM) works in the presence of the co-factor 2, 3-bisphosphoglycerate (2, 3-BPG). It has a molecular weight of 27 Kd, and are active as dimmers. The other type of the enzyme is the co-factor independent PGM (iPGM) has a molecular wight of 57 Kd. The enzyme is active in the monomer.
The distinction between the two forms of PGM can be determined by level of sensitivity to vanadate and metal ion requirement of iPGM. E.coli contains both iPGM and dPGM (Foster, 2010). Like the mammalian dPGMS, the E. coli dPGM forms a dimer which now enables researchers to analyze the sequence differences that are responsible for the variation in the quaternary structure.
The structure of the protein allows the function of the two enzyme domains to be assigned. One of the domains takes part in the phosphatase reaction, leading to the generation of the phosphoserine enzyme intermediate. The other domain participates in the phosphotransfterase reaction leading to reformation of the phosphoglycerate (Jedrzejas et al., (2000).
(EC 5.4.2.11 – Phosphoglycerate mutase, n.d)
Question 2
The enzyme phosphoglycerate mutase is involved in the glycolysis where it catalyzes the interconversion of 3-phosphoglycerate and 2-phosphoglycerate for zymomnas mobilis’ glyconeogenic and glycolytic pathways. The phospho group is transferred between the three phosphoglycerate carbon atoms. The co-factor dependent PGM (d PGM) catalyzes the transfer of a phosphoryl group between the co-factor and the monophosphoglycerate via an intermediate, phosphohistine. It is important in the transfer of a phospho group between monophosphoglycerates via a phosphoserine intermediate. The respective function of both iPGM and dPGM has not been established. However, research has shown that dPGM accounted for more activities (Foster, 2010).
The relevance of PH sensitivity in physiological processes because plays a role in regulating the activity of iPGM at different stages of an organism’s developmental cycle. For instance, the PH of a developing spore drops to about 6.5 during sporulation which leads to the reduction in the iPGM’s activities. The low PH allows 3PGA to accumulate and is stored in the dormant spore. This 3PGA is used to synthesis ATP required in spore germination, again leading to a sharp rise in the PH to 7.5-8. In non-spore forming organisms that are closely related to those that reproduce by sporulation, iPGMs, Mn2+ is also required for catalysis.
The mechanism of iPGM catalysis of 2-phosphoglycerate occurs in two steps as this is a phosphatase reaction in which a phosphate group is transferred from 2 or 3-phosphoglycerate,forming an enzyme-bound phosphoserine intermediate, then a phosphotransferase reaction. Meanwhile, the phosphate is transferred from the enzyme-bound phospherine back to the glycerate moiety.
| Reaction: |
2-phospho-D-glycerate = 3-phospho-D-glycerate. |
||||
|
|||||
(EC 5.4.2.11 – Phosphoglycerate mutase, n.d)
The co-factor dependent PGM is capable of catalyzing three various reactions, each with its own type of specificity. The reactions include the synthesis in which 1,3-DPGforms 2,3-DPG where 3-PGA is the primer, isomerisation reaction in which 3-phosphoglycerate (3-PGA) is formed from 2-phosphoglycerate (2-PGA) where the primer is 2,3-diphosphoglycerate (2,3-DPG) and degradation leading to formation of 3PGA from 2,3-DPG in a phosphatise activity. Also, the analysis of dPGM shows the enzyme ligand interaction which takes place to where the vanadates inhibit the mutase activity.
Phosphoglycerate mutases (PGMs) enzymes are important in glycolysis and gluconeogenesis reactions. In the glycolytic catalysis, 2-phosphoglycerate (2PG) is formed from 3-phosphoglycerate (3PG) through the intermediate 2, 3-bisphosphoglycerate. The reaction said to be energetically neutral since the Gibbs energy involved being approximately 1.1KJ/mol. The neutral state of the energy is absolutely necessary for the generation of the proper molecule required to continue the glycolysis process.
The reaction catalyzed by PGM
3PG + P-Enzyme → 2,3BPG + Enzyme → 2PG + P-Enzyme
3-phosphoglycerate -> intermediate -> 2-phosphoglycerate
Analyses of structure and sequence indicate that various families of the phosphatase enzymes contribute to different branches which are added to the parent enzyme. For example, histidine phosphatase superfamily is given this name because the catalytic reaction centres on the conserved on the His residue. This residue is transiently phosphorylated during the catalytic cycle. There are other conserved residues which interact with the phosphor group besides contributing to the “phosphate docket”. Different residues are added to the “phosphate pocket”. The families which contribute the residues differ in the three dimensions, position and sequence of a catalytically essential residue acidic residue Bond, White & Hunter, 2002.).
Question 3
- coli’s structure of dPGM is complexed with vanadate, which is a potent inhibitor. The presence of the inhibitor in the active site although there is evidence indicating that there is additional vanadate moieties at both or either ends. The presence of the inhibitor also represents a different binding mode from the one observed on the structural homologue proctatic acid phosphatase. The analysis of dPGM reveals a water molecule in the native E.coli dPGM structure. Once this molecule is activated by vanadate, the active site protein is likely to be dephosphorylated. The binding of substrates as well as other specific interactions of both forms of dPGM’s active and inactive forms may be easily studied with a provision of the forms’ high-resolution structure in conjunction with computational substrate molecule modelling in the active site (Jedrzejas et al. 2000).
iPGM differs from thedPGM in that it only catalyses out interconversion of 2PGA and 3PGA (1, 3, 4). The other difference is that catalysis by co-factor dependent phosphoglycerate mutase does not require metal ion while the iPGM absolutely or specifically require Mn 2+ ions hence making the enzyme’s activity exquisitely sensitive to PH (1, 4, 6-8). In clinical practice, a number of diagnostic procedures rely on the PH values to determine the cause or level of infection (Jedrzejas et al. 2000).
Fig:. DS-PAGE analysis of purification and protein expression: Lane 1, size markers; lane 2, total soluble protein for cells expressing dPGM; lane 3, purified dPGM; lane 4, total soluble protein for cells expressing iPGM; lane 5, purified iPGM (Fraser, Kvaratskhelia & White, 1999).
The E. coli PGM is a protein hence is soluble in water although to different extents depending on the provided conditions. One of the conditions is the presence of Isopryl-1-thio-β-D- galactopyranoside. iPGM’s solubility impacts its ability to aggregate with other groups through chemical reactions. Solubility infers that the protein has negative charges. The solubility discriminates the reactions the enzyme can catalyze (Fraser, Kvaratskhelia & White, 1999). As mutase, PGM is an important step during glycolysis. The transfer of a functional group such as a phosphor group from one position of a substrate to another is catalyzed by the enzyme, hence the reaction becomes isomerisation. It will not function if it not subjected to the right conditions for solubility to occur.
The co-factor dependent phosphoglycerate mutase (dPGM) is found in a dimeric active conformation in Escherichia coli (E.coli). It is reactions are based on the structural changes which occur on the histidine phosphorylation exhibits a number of features which are fundamental in the catalytic mechanisms of the enzyme. During reactions, the C-terminal 10-residue tail which does not appear in other structures is ordered well so that to interact well with other residues involved in substrate binding. During the displacement of a loop positioned close to or adjacent to the active histidine leads to the readjustment of previously overlooked residues into positions which will make it easy for them to directly influence the catalysis (Bond, White & Hunter, 2002.).
References
Foster, J., Davis, P., Raverdy, S., Sibley, M., Raleigh, E., Kumar, S., … Ahmed, N. (2010). Evolution of Bacterial Phosphoglycerate Mutases: Non-Homologous Isofunctional Enzymes Undergoing Gene Losses, Gains and Lateral Transfers. PLoS ONE, E13576-E13576.
Jedrzejas et al., (2000). Mechanism of Catalysis of the Cofactor-independent Phosphoglycerate Mutase from Bacillus stearothermophilus. CRYSTAL STRUCTURE OF THE COMPLEX WITH 2-PHOSPHOGLYCERATE. Journal of Biological Chemistry, 23146-23153.
Bond, C., White, M., & Hunter, W. (2002.). Mechanistic implications for Escherichia coli cofactor-dependent phosphoglycerate mutase based on the high-resolution crystal structure of a vanadate complex. Journal of Molecular Biology, 1071-1081.
Fraser, H., Kvaratskhelia, M., & White, M. (1999). The two analogous phosphoglycerate mutases of Escherichia coli. FEBS Letters, 455(3)344-348.
EC 5.4.2.11 – Phosphoglycerate mutase (2,3-diphosphoglycerate-dependent). (n.d.). Retrieved November 19, 2014, from http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/enzymes/GetPage.pl?ec_number=5.4.2.11
We can write this or a similar paper for you! Simply fill the order form!




